Update Apr, 20205: We have received inquiries about the infrastructure below, and while it generally represents our v1 architecture from 2022, we are currently on v3 of our backend and the diagram below no longer represents our full API architecture.
Background
Recent models, such as ESM-2 and AlphaFold2.0, have shown promise in helping to solve complex biotech challenges. Some have been deployed on HuggingFace, in GitHub repos, and more places online for experimentation. Yet, while we can now ween out unstable molecules early using predictions; leverage models like DNA-BERT for filtering out sequences with a high likelihood of splicing; and estimate a synthetic protein’s Tm, there remains a gap between the research and businesses trying to use it to solve their own challenges. BioLM helps connect businesses with biological modeling solutions, many of which require expertise in MLOps, MLEngineering, the biology, and of course data science, to leverage properly.
Organizations are moving away from their own internal ML teams, and are seeking out services like BioLM to jump-start their work. While larger companies can more easily maintain their own systems, BioLM offers a secure and scalable solution for businesses looking to get that head start without all the overhead. It takes time, effort, and cost to specialize and deploy these algorithms for purpose. Containerization becomes critical for managing conflicting dependencies – again, more time. BioLM APIs abstract away these problems and deliver results via simple REST requests. We (and you) can then consume those APIs to create incredible API-based systems and scientific applications. Here’s how it’s done.
Scalable Infrastructure
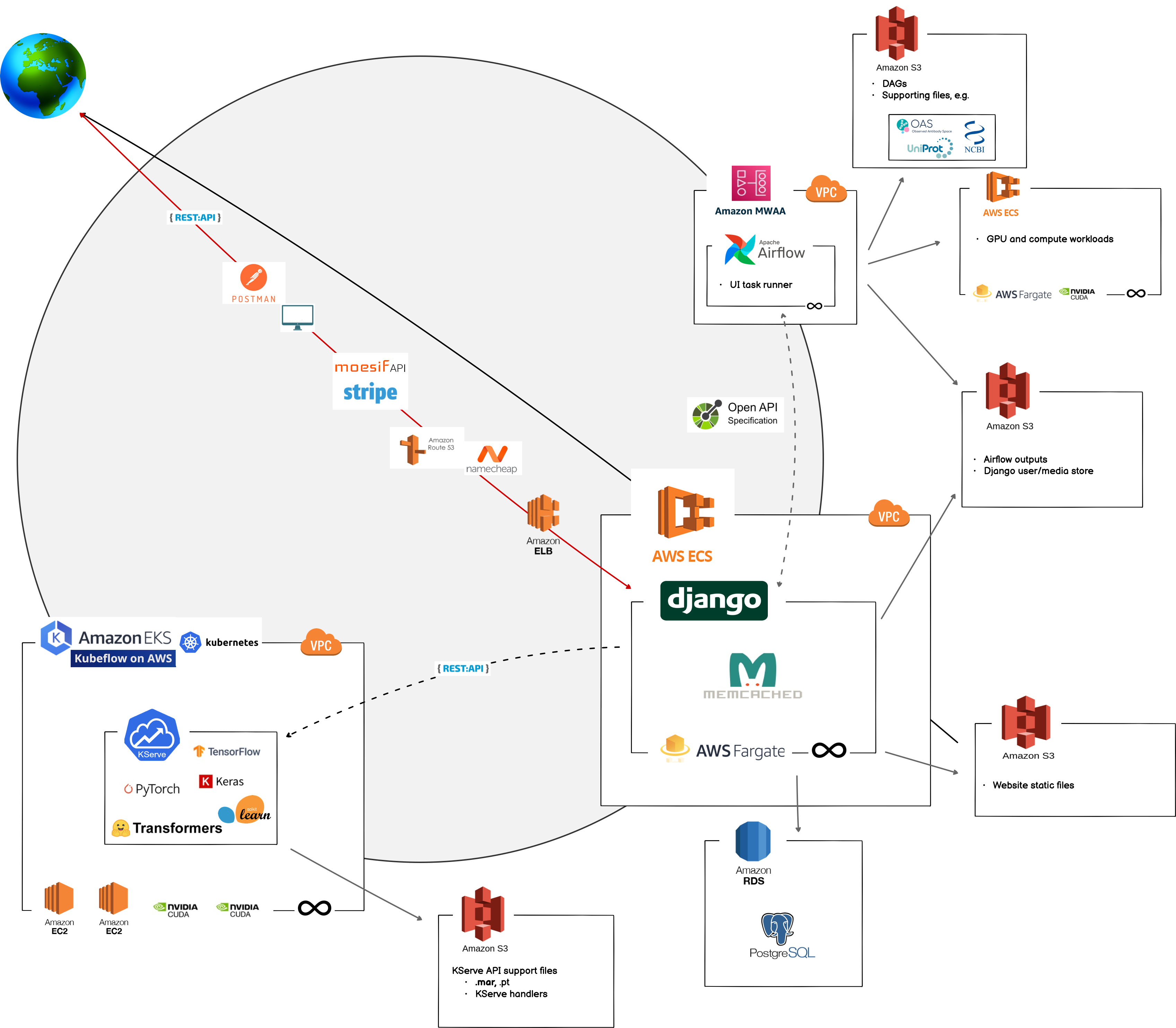
BioLM’s infrastructure is distributed across multiple services, primarily with serverless platforms that can access GPUs and spot cloud-instances. An async Django web-app functions as the primary ingress point for users. All pipelines triggered via UIs and all API calls route through here. This first touch-point is 100% scalable through AWS’ HTTP load balancer and AWS scalable ECS container services. Zero request per second? Zero cost. Thousands of requests per second? Containers can instantly scale up in real-time.
Moesif monitors consumption of APIs and pipelines and pro-rates them, linking with billing systems. The team there has been great, and I’ve really appreciated their help. Ultimately, that data is sent to Stripe, where billing and invoices are securely handled. Whether users are on a fixed-rate or pro-rated plan, Stripe handles payments.
APIs & Web Apps
Outside of Django live two other critical systems: Airflow and Kubernetes on AWS. Many of our models are deployed via Kubeflow model serving or Ray, where we can take advantage of PyTorch’s Torchserve, KNative serving for scikit-learn or other CPU-bound operations, Nvidia Triton server for Tensorflow and multiple-model-per-GPU serving. With Kubernetes and real-time monitoring, the number of pods, nodes – in effect the model serving – can scale up and down seamlessly in response to requests. Spot instances are used whenever possible. All of this allows keeping costs low for end users, without sacrificing any power, performance, or results. Each model’s GPU, CPU, and memory requirements are specified on deployment and then granted to the instance when the model is loaded into memory on the server.
Un-coincidentally, this stack and MWAA Airflow are essentially serverless, too, leaving only the databases running when there is no load (now being transitioned to serverless Neon DB). Airflow needs no introduction to many people. It handles longer-running tasks and backs the UI applications. If we want to rent a GPU for longer-term compute tasks, we can do that here, too. UI results are stored in encrypted user-folders in S3 so you have access to them. On the other hand, we do not store API results. While we are very careful securing sequences in transmission and at rest, we are really not interested in keeping them around on our servers, and happily remove them when possible.
Discussion
So where does all this leave us now? Five years ago we were building models, packaging them in Python, and training them in the cloud for long periods of time before we could even touch them. Now we can use massive genomic/proteomic information and published research to deliver better predictions in a fraction of the time. Real-time chatbots, recommendation systems, ride-share pickups, even news reels are powered by infrastructure like this that make more real-time analysis possible. Don’t get me wrong, the infrastructure is not the solution, but it’s a good start to innovation. The outcomes must be paired with an understanding of how to use them – the domain expertise.
I’m sure many people downloaded and tried ESMFold for Kaggle’s Tm competition, but weren’t sure what to do with the output files to make a Tm prediction. This, judging by the large gap between the top two or three 0.75+ Spearman scores and the rest of the thousands of models performing at < 0.69 or worse. Strictly tech-companies with no biological expertise who have deployed these models are disillusioned in this way, along with their customers, who are looking like deer in the headlights at how to use the expensive solution they just paid for and no hand to hold. “You can use this database; this best model.” It doesn’t work that way. The dots need connection, and if your solutions provider has no experience modeling DNA or AAs, they won’t be able to help you much beyond simple deployments.
Conclusion
Ginkgo’s launch of Enzyme Services earlier this week is a great example of what’s possible when you pair high-throughput modeling and screening of molecules with synthesis. I imagine some of these technologies I mentioned, from Django to the model containerization, are used over there. Since Ginkgo already has to maintain existing web and other technology stacks for their core business, the opportunity cost to deploy this system in-house is relatively low for them; and the model outputs feed directly into their biofactory business. Things might not be as clear for other companies whose core competency is in other areas. Which is why I want to bring these types of capabilities to more businesses, maybe like yours. Need to classify, regress, or generate DNA or proteins? Need to scale it? BioLM services might be able to help.